GNXASCurrent versions: 12.2023/11.2013/12.2006
Info and NewsLatest GNXAS intensive courses: Camerino, Italy (2009), Melbourne, Australia (2011), Riberao Preto, Brasil (2013), Karlsruhe, Germany (Aug 2015), Rennes, France (June 2016), Krakow, Poland (August 2018), Nagoya, Japan (August 2023). Latest news and info about the programs
INTRODUCTION to GNXASThe GNXAS package is an advanced software for EXAFS data analysis based on multiple-scattering (MS) calculations and a rigorous fitting procedure of the raw experimental data. The main characteristic of the software are:
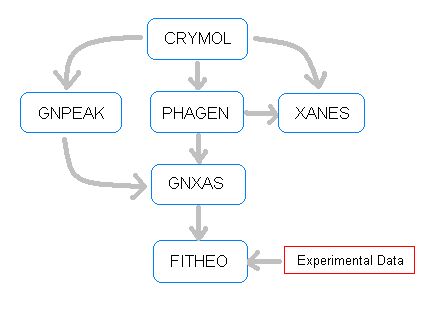
Its flexibility and advanced characteristics are essential to perform a modern EXAFS data-analysis and to extract accurately all the valuable information contained in your data. GNXAS was supported by the Italian Consiglio Nazionale delle Ricerche (CNR) (last contract 98.00475.ST76). Later on, INFM supported the developments in 1999-2003. PROGRAM DESCRIPTIONThe backbone of the GNXAS package is composed of three main codes, in the logical sequence they are:
Two other programs are able to prepare automatically the input for PHAGEN and GNXAS starting directly from model molecular position or crystallographic data. These are particularly useful in the case of complex structures where the configuration counting is not trivial. These programs are:
Several other programs were written to facilitate the output readout, in particular:
Three additional programs have been written to facilitate the analysis of disordered systems (including liquids) where the structure is described in terms of radial distribution functions g(r). These are:
Finally the GNXAS distribution includes miscellaneous programs useful for XAS based research:
The extended GNXAS package includes also several other advanced or utility Linux programs that are available to users upon request, described in the recent Full and updated GNXAS documentation (2009): the GNXAS 'red-book'.
DOCUMENTATION
WHO WROTE GNXASSeveral scientists have contributed to the development of GNXAS since 1989. It contains the most up to date algorithms for EXAFS data analysis directly coded by the investigators. It was soon realized that the use of these programs could have been appreciated by scientists in various fields willing to perform advanced applications of the EXAFS technique. For this reason the programs were made available to the scientific community. We should warn the users however that as a result of this history GNXAS is not a trivial and "user proof" code. The complexity of some of the algorithms makes the required input very cumbersome and INPUT mistakes can be sometimes hard to identify. The research effort was based on the work of C. R. Natoli (INFN - Laboratori Nazionali di Frascati), a pioneer of the multiple-scattering applications to the EXAFS spectroscopy. Natoli is the main author of the PHAGEN program where all the theory for the potential calculation is included. The GNXAS code was mainly written by A. Filipponi - Universita' dell'Aquila, implementing efficient codes for MS calculations with fast matrix products and continued fraction approaches. FITHEO has been mainly written by A. Di Cicco Universita' di Camerino. A complete version of GNXAS was already available in 1990. Several algorithms and approaches implemented in the package have been jointly developed by the authors. Other programs and algorithms, like RMC-GNXAS, EDXRD (Di Cicco), and DECONV, XASAM (Filipponi) have been developed separately by the authors. Other colleagues participated to major theoretical developments and/or have partly contributed to the development of the package. Among them we acknowledge, in chronological order:
WHERE TO FIND GNXASDistribution of the code is provided by the XAS group @ Camerino University working at the Sezione di Fisica . The executable codes are made available to users without charge provided that they are used for NO PROFIT scientific research and full acknowledgement is given to the authors. GNXAS must NOT be used for military research, nuclear research, or any kind of classified research. The authors decline any responsibility for erroneous results that can be obtained as a consequence of proper or improper use of the package. The executable are presently available for the following operative systems: The executables can be downloaded following our registration procedures. Last revised 20 Aug 2024 - Andrea Di Cicco |